
Technology review
In recent years, a growing number of key players have entered the field of genome editing, driven by the transformative potential of these technologies to modify genetic material and treat a broad spectrum of previously untreatable diseases. One well-established method involves introducing double-strand breaks (DSBs) in DNA, a strategy pioneered with tools such as meganucleases, zinc finger nucleases, and transcription activator-like effector nucleases. More recently, CRISPR-Cas systems have revolutionised genome editing by offering unprecedented levels of precision, efficiency, and accessibility.
However, a newer and more powerful genome-editing tool that offers a promising alternative by converting a single base (pair) without the need of DSBs while using the natural host cell DSB repair mechanism is ‘base editing’. First described by David Liu’s lab (Komor et al., 2016), base editors (BEs) are built upon CRISPR-Cas systems and can be categorised into two main systems: cytosine base editors (CBEs) and adenine base editors (ABEs).
Typically, CBEs are engineered by fusing a cytosine deaminase like APOBEC to a deactivated or ’dead’ Cas9 nickase. The deaminase converts cytosine (C) to uracil (U) and during DNA replication, uracil is interpreted as thymine (T) by the cellular machinery, resulting in a permanent C-G to T-A substitution. Similarly, ABEs are created by fusing an adenosine deaminase to a Cas9 variant. ABEs subsequently convert adenosine (A) to inosine (I) in the targeted DNA strand, which is read as guanine (G) during replication, leading to a A-T to G-C substitution.
Both base editing systems can exhibit off-target effects, though these are generally less frequent compared to those observed with traditional CRISPR-Cas9 methods. Efficiency can also vary depending on factors such as the context of the target sequence as well as cell type. Current CBEs are limited to converting C-G to T-A pairs, while ABEs are restricted to A-T to G-C conversions. However, recent advancements include new CBE compositions that use adenine deaminases and Cas9 proteins to enable conversion to other bases such as adenosine, guanosine, and thymidine (e.g. EP4006157).
Indications treated
Particularly in the context of precision and personalised medicine, base editors are increasingly recognised as powerful tools for advancing biomedical research and enabling the treatment and prevention of complex genetic diseases. As of May 2025, key players include The Broad Institute, Verve Therapeutics, Beam Therapeutics, Bioray Laboratories, and Great Ormond Street Hospital, London, UK.
In July 2022, Verve Therapeutics announced that the first patient had been dosed with VERVE-101, a treatment for heterozygous familial hypercholesterolemia (HeFH) (NCT05398029). VERVE-101 utilises base editing technology to permanently knock out the PCSK9 gene in the liver by introducing a point mutation from A-T to G-C in the PCSK9 gene. This creates a premature stop codon, which leads to mRNA degradation and a reduction in circulating cholesterol levels.
Base editors may also be used to restore the wild-type function of a protein. Beam Therapeutics announced enrolment for its BEACON trial for the treatment of sickle cell disease (SCD). BEAM-101 is intended to treat all three forms of SCD which carry a specific T-to-A mutation in the β-globin protein, resulting in sticky β-globin proteins.
In December 2022, it was reported that a teenager in the UK is in complete remission from resistant T-cell acute lymphoblastic leukaemia following treatment with a novel base-edited chimeric antigen receptor T cell therapy at Great Ormond Street Hospital, London, UK.
More recently, BEs were reported as an approach in treating cystic fibrosis (Roberts, 2025). Accordingly, this technology holds potential for clinical expansion to address other genetic diseases with unmet medical needs.
Patent considerations
Like any other patentable invention under the EPC, base editor inventions must fulfil the basic patentability requirements and must not be excluded from patentability. Suitable patent claims may be directed to specific sequences, expression constructs and delivery systems, methods and kits, or compositions and their medical uses.
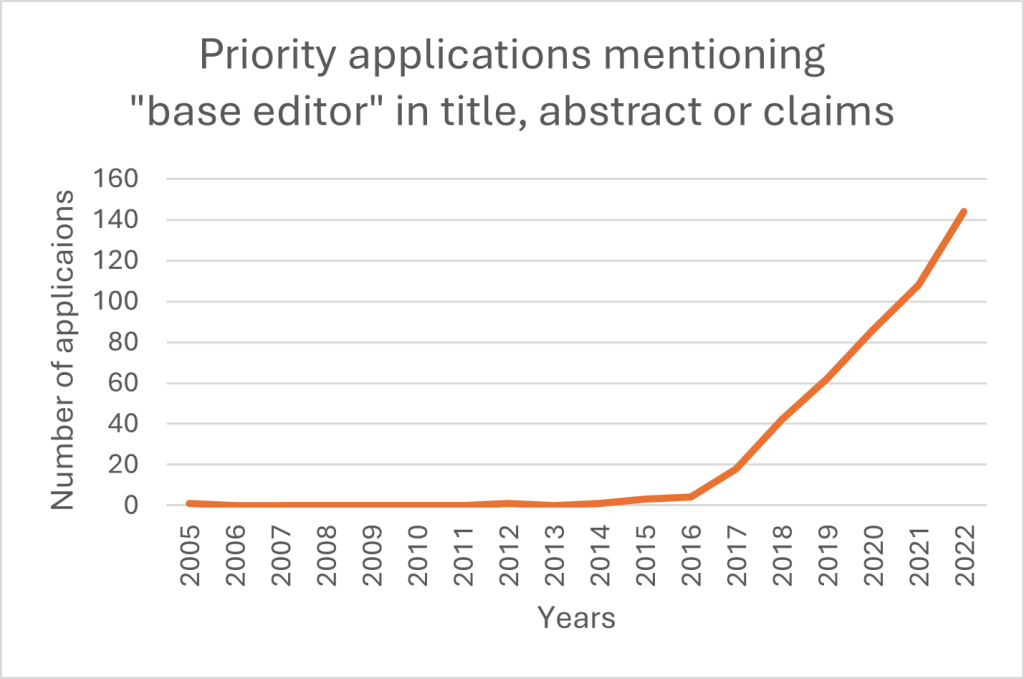
Yet, patent considerations are quite complex due to the competitive and rapidly evolving nature of the field. As figure 1 below shows, priority patent applications filed worldwide mentioning ‘base editor’ have been increasing consistently over the past nine years.
As the technology becomes more established, patent claims still remain relatively broad, often covering fusion proteins that combine a deaminase with a Cas protein. However, since BE systems typically involve sequence-specific guide RNAs (gRNAs) and the field is becoming increasingly competitive, it is advisable to include specific sequences and/or chemical modification patterns of the gRNAs. In particular, the pattern of chemical modifications in a specific gRNA is critical in assessing inventive step, especially in terms of enhanced stability, specificity, and efficiency compared to prior art.
Further, patent claims need to be clear. As illustrated by EP3592777, a “nucleic acid programmable DNA binding protein” may be objected to for lack of clarity. Also, wording in relation to ‘variants’ may have to be deleted to address possible sufficiency objections. Also, since BEs aim to modify the genetic material, disclaimers may be incorporated specifying that the method is not a method for modifying the germ line of humans. As the field is relatively new the jurisprudence is still in its early stages at the EPO. We are expecting a lot of EPO oppositions in this area in the years to come, which will develop the case law accordingly.
Developing effective delivery methods to target base editors to the desired tissues or cell types in vivo is essential for their clinical translation. Existing technology in this area includes tagging RNA with Cell-Penetrating Peptides or N-acetylgalactosamine, encapsulating RNA in nanoparticles or viral vectors, such as AAV, which can be used to deliver RNA molecules into both dividing and non-dividing cells with long-lasting expression. We expect this particular aspect to be the subject of intense competition as companies seek to develop platform technologies that will allow rapid development of medicines that can be economically targeted to individuals or small patient cohorts, essential for tackling rare disease.
More recently, artificial intelligence (AI) has been taking on an increased role in identifying and designing novel gene editors that are functional and that show comparable or improved genome-editing activity and specificity relative to naturally occurring gene editors. Accordingly, as AI inventions are on the rise, the EPO has responded to the emergence of AI in patent applications by clarifying the scope of Article 52 EPC to also include AI inventions and refining its guidelines (EPO GL G-II, 3.3.1).
Summary
Base editors are a powerful and precise gene-editing tool that enable specific DNA changes with high accuracy, without causing double-strand breaks. This reduces the risk of unwanted genetic alterations, making base editing a potentially safer and more effective option for introducing single point mutations. When considering patenting this technology, it is crucial to consider factors such as target specificity and sequence, type of base conversion, and delivery methods.
The biotechnology group at GJE has particular experience in patenting RNA technologies, including base editing. Our AI patent team also advises clients who are using machine learning platforms for developing biotechnological innovations. We advise both innovative biotech companies and investors across Europe, the US and Asia. To discuss your biotech IP strategy, please contact us at biotech@gje.com.
References
Komor et al. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 533; 420-424. Available online.
EP4006157; available online.
EP3592777; available online.
VERVE-101 Study; available online.
BEACON Trial; available online.
Roberts, R. 2025. An Innovative Approach to Treating Cystic Fibrosis Using Base Editors. Available online.
World-first use of base-edited CAR T-cells to treat resistant leukaemia | UCL News – UCL – University College London; accessed May 2025. Available here.


